标签:== closed spi matrix event ref ret pac 技术
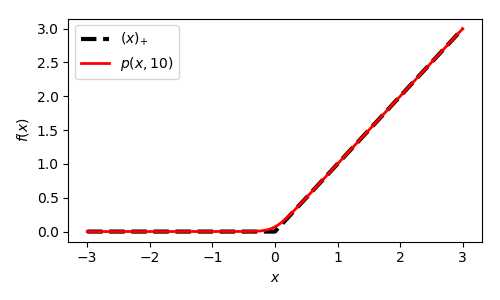 下面给出p-function的1阶及2阶导数:
下面给出p-function的1阶及2阶导数: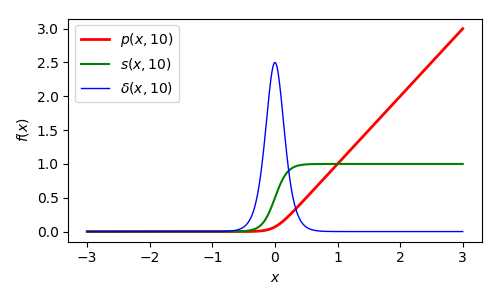 Part Ⅱ
Part Ⅱ
1 # Smooth Support Vector Machine之实现 2 3 import numpy 4 from matplotlib import pyplot as plt 5 6 7 def spiral_point(val, center=(0, 0)): 8 rn = 0.4 * (105 - val) / 104 9 an = numpy.pi * (val - 1) / 25 10 11 x0 = center[0] + rn * numpy.sin(an) 12 y0 = center[1] + rn * numpy.cos(an) 13 z0 = -1 14 x1 = center[0] - rn * numpy.sin(an) 15 y1 = center[1] - rn * numpy.cos(an) 16 z1 = 1 17 18 return (x0, y0, z0), (x1, y1, z1) 19 20 21 def spiral_data(valList): 22 dataList = list(spiral_point(val) for val in valList) 23 data0 = numpy.array(list(item[0] for item in dataList)) 24 data1 = numpy.array(list(item[1] for item in dataList)) 25 return data0, data1 26 27 28 # 生成训练数据集 29 trainingValList = numpy.arange(1, 101, 1) 30 trainingData0, trainingData1 = spiral_data(trainingValList) 31 trainingSet = numpy.vstack((trainingData0, trainingData1)) 32 # 生成测试数据集 33 testValList = numpy.arange(1.5, 101.5, 1) 34 testData0, testData1 = spiral_data(testValList) 35 testSet = numpy.vstack((testData0, testData1)) 36 37 38 class SSVM(object): 39 40 def __init__(self, trainingSet, c=1, mu=1, beta=100): 41 self.__trainingSet = trainingSet # 训练集数据 42 self.__c = c # 误差项权重 43 self.__mu = mu # gaussian kernel参数 44 self.__beta = beta # 光滑化参数 45 46 self.__A, self.__D = self.__get_AD() 47 48 49 def get_cls(self, x, alpha, b): 50 A, D = self.__A, self.__D 51 mu = self.__mu 52 53 x = numpy.array(x).reshape((-1, 1)) 54 KAx = self.__get_KAx(A, x, mu) 55 clsVal = self.__calc_hVal(KAx, D, alpha, b) 56 if clsVal > 0: 57 return 1 58 elif clsVal == 0: 59 return 0 60 else: 61 return -1 62 63 64 def get_accuracy(self, dataSet, alpha, b): 65 ‘‘‘ 66 正确率计算 67 ‘‘‘ 68 rightCnt = 0 69 for row in dataSet: 70 clsVal = self.get_cls(row[:2], alpha, b) 71 if clsVal == row[2]: 72 rightCnt += 1 73 accuracy = rightCnt / dataSet.shape[0] 74 return accuracy 75 76 77 def optimize(self, maxIter=100, epsilon=1.e-9): 78 ‘‘‘ 79 maxIter: 最大迭代次数 80 epsilon: 收敛判据, 梯度趋于0则收敛 81 ‘‘‘ 82 A, D = self.__A, self.__D 83 c = self.__c 84 mu = self.__mu 85 beta = self.__beta 86 87 alpha, b = self.__init_alpha_b((A.shape[1], 1)) 88 KAA = self.__get_KAA(A, mu) 89 90 JVal = self.__calc_JVal(KAA, D, c, beta, alpha, b) 91 grad = self.__calc_grad(KAA, D, c, beta, alpha, b) 92 Hess = self.__calc_Hess(KAA, D, c, beta, alpha, b) 93 94 for i in range(maxIter): 95 if self.__converged1(grad, epsilon): 96 return alpha, b, True 97 98 dCurr = -numpy.matmul(numpy.linalg.inv(Hess), grad) 99 ALPHA = self.__calc_ALPHA_by_ArmijoRule(alpha, b, JVal, grad, dCurr, KAA, D, c, beta) 100 101 delta = ALPHA * dCurr 102 alphaNew = alpha + delta[:-1, :] 103 bNew = b + delta[-1, -1] 104 JValNew = self.__calc_JVal(KAA, D, c, beta, alphaNew, bNew) 105 if self.__converged2(delta, JValNew - JVal, epsilon): 106 return alphaNew, bNew, True 107 108 alpha, b, JVal = alphaNew, bNew, JValNew 109 grad = self.__calc_grad(KAA, D, c, beta, alpha, b) 110 Hess = self.__calc_Hess(KAA, D, c, beta, alpha, b) 111 else: 112 if self.__converged1(grad, epsilon): 113 return alpha, b, True 114 return alpha, b, False 115 116 117 def __converged2(self, delta, JValDelta, epsilon): 118 val1 = numpy.linalg.norm(delta) 119 val2 = numpy.linalg.norm(JValDelta) 120 if val1 <= epsilon or val2 <= epsilon: 121 return True 122 return False 123 124 125 def __calc_ALPHA_by_ArmijoRule(self, alphaCurr, bCurr, JCurr, gCurr, dCurr, KAA, D, c, beta, C=1.e-4, v=0.5): 126 i = 0 127 ALPHA = v ** i 128 delta = ALPHA * dCurr 129 alphaNext = alphaCurr + delta[:-1, :] 130 bNext = bCurr + delta[-1, -1] 131 JNext = self.__calc_JVal(KAA, D, c, beta, alphaNext, bNext) 132 while True: 133 if JNext <= JCurr + C * ALPHA * numpy.matmul(dCurr.T, gCurr)[0, 0]: break 134 i += 1 135 ALPHA = v ** i 136 delta = ALPHA * dCurr 137 alphaNext = alphaCurr + delta[:-1, :] 138 bNext = bCurr + delta[-1, -1] 139 JNext = self.__calc_JVal(KAA, D, c, beta, alphaNext, bNext) 140 return ALPHA 141 142 143 def __converged1(self, grad, epsilon): 144 if numpy.linalg.norm(grad) <= epsilon: 145 return True 146 return False 147 148 149 def __p(self, x, beta): 150 val = numpy.log(numpy.exp(beta * x) + 1) / beta 151 return val 152 153 154 def __s(self, x, beta): 155 val = 1 / (numpy.exp(-beta * x) + 1) 156 return val 157 158 159 def __d(self, x, beta): 160 term = numpy.exp(beta * x) 161 val = beta * term / (term + 1) ** 2 162 return val 163 164 165 def __calc_Hess(self, KAA, D, c, beta, alpha, b): 166 Hess_J1 = self.__calc_Hess_J1(alpha) 167 Hess_J2 = self.__calc_Hess_J2(KAA, D, c, beta, alpha, b) 168 Hess = Hess_J1 + Hess_J2 169 return Hess 170 171 172 def __calc_Hess_J2(self, KAA, D, c, beta, alpha, b): 173 Hess_J2 = numpy.zeros((KAA.shape[0] + 1, KAA.shape[0] + 1)) 174 Y = numpy.matmul(D, numpy.ones((D.shape[0], 1))) 175 YY = numpy.matmul(Y, Y.T) 176 KAAYY = KAA * YY 177 178 z = 1 - numpy.matmul(KAAYY, alpha) - Y * b 179 p = numpy.array(list(self.__p(z[i, 0], beta) for i in range(z.shape[0]))).reshape((-1, 1)) 180 s = numpy.array(list(self.__s(z[i, 0], beta) for i in range(z.shape[0]))).reshape((-1, 1)) 181 d = numpy.array(list(self.__d(z[i, 0], beta) for i in range(z.shape[0]))).reshape((-1, 1)) 182 term = s * s + p * d 183 184 for k in range(Hess_J2.shape[0] - 1): 185 for l in range(k + 1): 186 val = c * Y[k, 0] * Y[l, 0] * numpy.sum(KAA[:, k:k+1] * KAA[:, l:l+1] * term) 187 Hess_J2[k, l] = Hess_J2[l, k] = val 188 val = c * Y[k, 0] * numpy.sum(KAA[:, k:k+1] * term) 189 Hess_J2[k, -1] = Hess_J2[-1, k] = val 190 val = c * numpy.sum(term) 191 Hess_J2[-1, -1] = val 192 return Hess_J2 193 194 195 def __calc_Hess_J1(self, alpha): 196 I = numpy.identity(alpha.shape[0]) 197 term = numpy.hstack((I, numpy.zeros((I.shape[0], 1)))) 198 Hess_J1 = numpy.vstack((term, numpy.zeros((1, term.shape[1])))) 199 return Hess_J1 200 201 202 def __calc_grad(self, KAA, D, c, beta, alpha, b): 203 grad_J1 = self.__calc_grad_J1(alpha) 204 grad_J2 = self.__calc_grad_J2(KAA, D, c, beta, alpha, b) 205 grad = grad_J1 + grad_J2 206 return grad 207 208 209 def __calc_grad_J2(self, KAA, D, c, beta, alpha, b): 210 grad_J2 = numpy.zeros((KAA.shape[0] + 1, 1)) 211 Y = numpy.matmul(D, numpy.ones((D.shape[0], 1))) 212 YY = numpy.matmul(Y, Y.T) 213 KAAYY = KAA * YY 214 215 z = 1 - numpy.matmul(KAAYY, alpha) - Y * b 216 p = numpy.array(list(self.__p(z[i, 0], beta) for i in range(z.shape[0]))).reshape((-1, 1)) 217 s = numpy.array(list(self.__s(z[i, 0], beta) for i in range(z.shape[0]))).reshape((-1, 1)) 218 term = p * s 219 220 for k in range(grad_J2.shape[0] - 1): 221 val = -c * Y[k, 0] * numpy.sum(Y * KAA[:, k:k+1] * term) 222 grad_J2[k, 0] = val 223 grad_J2[-1, 0] = -c * numpy.sum(Y * term) 224 return grad_J2 225 226 227 def __calc_grad_J1(self, alpha): 228 grad_J1 = numpy.vstack((alpha, [[0]])) 229 return grad_J1 230 231 232 def __calc_JVal(self, KAA, D, c, beta, alpha, b): 233 J1 = self.__calc_J1(alpha) 234 J2 = self.__calc_J2(KAA, D, c, beta, alpha, b) 235 JVal = J1 + J2 236 return JVal 237 238 239 def __calc_J2(self, KAA, D, c, beta, alpha, b): 240 tmpOne = numpy.ones((KAA.shape[0], 1)) 241 x = tmpOne - numpy.matmul(numpy.matmul(numpy.matmul(D, KAA), D), alpha) - numpy.matmul(D, tmpOne) * b 242 p = numpy.array(list(self.__p(x[i, 0], beta) for i in range(x.shape[0]))) 243 J2 = numpy.sum(p * p) * c / 2 244 return J2 245 246 247 def __calc_J1(self, alpha): 248 J1 = numpy.sum(alpha * alpha) / 2 249 return J1 250 251 252 def __get_KAA(self, A, mu): 253 KAA = numpy.zeros((A.shape[1], A.shape[1])) 254 for rowIdx in range(KAA.shape[0]): 255 for colIdx in range(rowIdx + 1): 256 x1 = A[:, rowIdx:rowIdx+1] 257 x2 = A[:, colIdx:colIdx+1] 258 val = self.__calc_gaussian(x1, x2, mu) 259 KAA[rowIdx, colIdx] = KAA[colIdx, rowIdx] = val 260 return KAA 261 262 263 def __init_alpha_b(self, shape): 264 ‘‘‘ 265 alpha, b之初始化 266 ‘‘‘ 267 alpha, b = numpy.zeros(shape), 0 268 return alpha, b 269 270 271 def __get_KAx(self, A, x, mu): 272 KAx = numpy.zeros((A.shape[1], 1)) 273 for rowIdx in range(KAx.shape[0]): 274 x1 = A[:, rowIdx:rowIdx+1] 275 val = self.__calc_gaussian(x1, x, mu) 276 KAx[rowIdx, 0] = val 277 return KAx 278 279 280 def __calc_hVal(self, KAx, D, alpha, b): 281 hVal = numpy.matmul(numpy.matmul(alpha.T, D), KAx)[0, 0] + b 282 return hVal 283 284 285 def __calc_gaussian(self, x1, x2, mu): 286 val = numpy.exp(-mu * numpy.linalg.norm(x1 - x2) ** 2) 287 # val = numpy.sum(x1 * x2) 288 return val 289 290 291 def __get_AD(self): 292 A = self.__trainingSet[:, :2].T 293 D = numpy.diag(self.__trainingSet[:, 2]) 294 return A, D 295 296 297 class SpiralPlot(object): 298 299 def spiral_data_plot(self, trainingData0, trainingData1, testData0, testData1): 300 fig = plt.figure(figsize=(5, 5)) 301 ax1 = plt.subplot() 302 ax1.scatter(trainingData1[:, 0], trainingData1[:, 1], c="red", marker="o", s=10, label="training data - Positive") 303 ax1.scatter(trainingData0[:, 0], trainingData0[:, 1], c="blue", marker="o", s=10, label="training data - Negative") 304 ax1.scatter(testData1[:, 0], testData1[:, 1], c="red", marker="x", s=10, label="test data - Positive") 305 ax1.scatter(testData0[:, 0], testData0[:, 1], c="blue", marker="x", s=10, label="test data - Negative") 306 ax1.set(xlim=(-0.5, 0.5), ylim=(-0.5, 0.5), xlabel="$x_1$", ylabel="$x_2$") 307 plt.legend(fontsize="x-small") 308 fig.tight_layout() 309 fig.savefig("spiral.png", dpi=100) 310 plt.close() 311 312 313 def spiral_pred_plot(self, trainingData0, trainingData1, testData0, testData1, ssvmObj, alpha, b): 314 x = numpy.linspace(-0.5, 0.5, 500) 315 y = numpy.linspace(-0.5, 0.5, 500) 316 x, y = numpy.meshgrid(x, y) 317 z = numpy.zeros(shape=x.shape) 318 for rowIdx in range(x.shape[0]): 319 for colIdx in range(x.shape[1]): 320 z[rowIdx, colIdx] = ssvmObj.get_cls((x[rowIdx, colIdx], y[rowIdx, colIdx]), alpha, b) 321 cls2color = {-1: "blue", 0: "white", 1: "red"} 322 323 fig = plt.figure(figsize=(5, 5)) 324 ax1 = plt.subplot() 325 ax1.contourf(x, y, z, levels=[-1.5, -0.5, 0.5, 1.5], colors=["blue", "white", "red"], alpha=0.3) 326 ax1.scatter(trainingData1[:, 0], trainingData1[:, 1], c="red", marker="o", s=10, label="training data - Positive") 327 ax1.scatter(trainingData0[:, 0], trainingData0[:, 1], c="blue", marker="o", s=10, label="training data - Negative") 328 ax1.scatter(testData1[:, 0], testData1[:, 1], c="red", marker="x", s=10, label="test data - Positive") 329 ax1.scatter(testData0[:, 0], testData0[:, 1], c="blue", marker="x", s=10, label="test data - Negative") 330 ax1.set(xlim=(-0.5, 0.5), ylim=(-0.5, 0.5), xlabel="$x_1$", ylabel="$x_2$") 331 plt.legend(loc="upper left", fontsize="x-small") 332 fig.tight_layout() 333 fig.savefig("pred.png", dpi=100) 334 plt.close() 335 336 337 338 if __name__ == "__main__": 339 ssvmObj = SSVM(trainingSet, c=0.1, mu=250, beta=100) 340 alpha, b, tab = ssvmObj.optimize() 341 accuracy1 = ssvmObj.get_accuracy(trainingSet, alpha, b) 342 print("Accuracy on trainingSet is {}%".format(accuracy1 * 100)) 343 accuracy2 = ssvmObj.get_accuracy(testSet, alpha, b) 344 print("Accuracy on testSet is {}%".format(accuracy2 * 100)) 345 346 spObj = SpiralPlot() 347 spObj.spiral_data_plot(trainingData0, trainingData1, testData0, testData1) 348 spObj.spiral_pred_plot(trainingData0, trainingData1, testData0, testData1, ssvmObj, alpha, b)
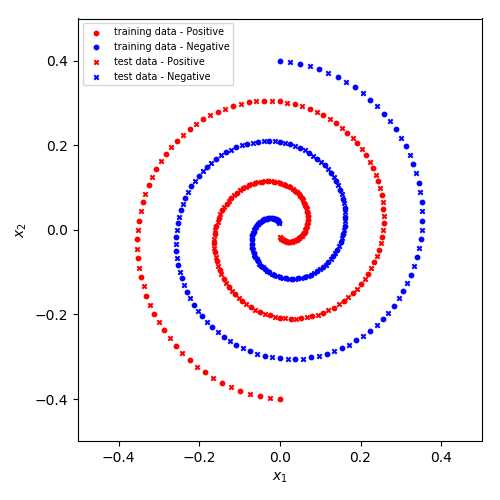 很显然, 此数据集非线性可分.
很显然, 此数据集非线性可分.
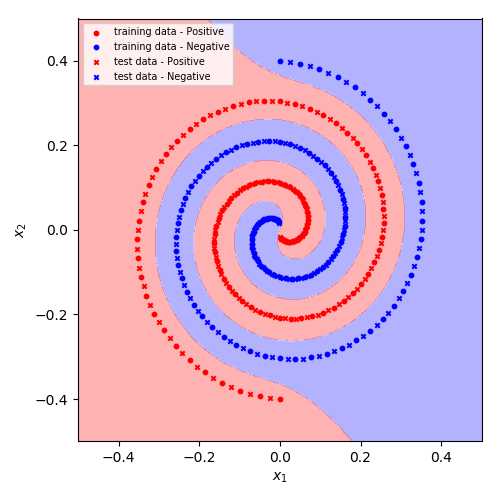 此模型在训练集、测试集上的准确率均达到100%.
此模型在训练集、测试集上的准确率均达到100%.Smooth Support Vector Machine - Python实现
标签:== closed spi matrix event ref ret pac 技术
原文地址:https://www.cnblogs.com/xxhbdk/p/12275567.html