标签:
根据决策值和真实标签画ROC曲线,同时计算AUC的值
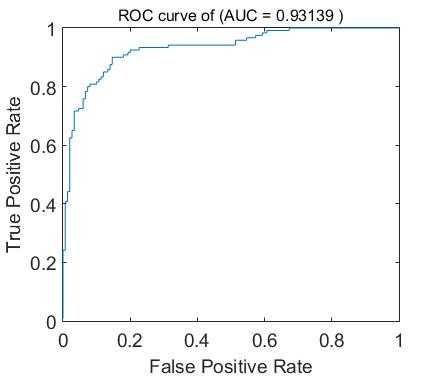
function auc = roc_curve(deci,label_y) %%deci=wx+b, label_y, true label
[val,ind] = sort(deci,‘descend‘);
roc_y = label_y(ind);
stack_x = cumsum(roc_y == -1)/sum(roc_y == -1);
stack_y = cumsum(roc_y == 1)/sum(roc_y == 1);
auc = sum((stack_x(2:length(roc_y),1)-stack_x(1:length(roc_y)-1,1)).*stack_y(2:length(roc_y),1))
%Comment the above lines if using perfcurve of statistics toolbox
%[stack_x,stack_y,thre,auc]=perfcurve(label_y,deci,1);
plot(stack_x,stack_y);
xlabel(‘False Positive Rate‘);
ylabel(‘True Positive Rate‘);
title([‘ROC curve of (AUC = ‘ num2str(auc) ‘ )‘]);
end
代码来自林智仁网站:https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/#roc_curve_for_binary_svm
function auc = plotroc(y,x,params)
%plotroc draws the recevier operating characteristic(ROC) curve.
%
%auc = plotroc(training_label, training_instance [, libsvm_options -v cv_fold])
% Use cross-validation on training data to get decision values and plot ROC curve.
%
%auc = plotroc(testing_label, testing_instance, model)
% Use the given model to predict testing data and obtain decision values
% for ROC
%
% Example:
%
% load(‘heart_scale.mat‘);
% plotroc(heart_scale_label, heart_scale_inst,‘-v 5‘);
%
% [y,x] = libsvmread(‘heart_scale‘);
% model = svmtrain(y,x);
% plotroc(y,x,model);
rand(‘state‘,0); % reset random seed
if nargin < 2
help plotroc
return
elseif isempty(y) | isempty(x)
error(‘Input data is empty‘);
elseif sum(y == 1) + sum(y == -1) ~= length(y)
error(‘ROC is only applicable to binary classes with labels 1, -1‘); % check the trainig_file is binary
elseif exist(‘params‘) && ~ischar(params)
model = params;
[predict_label,mse,deci] = svmpredict(y,x,model) ;% the procedure for predicting
auc = roc_curve(deci*model.Label(1),y);
else
if ~exist(‘params‘)
params = [];
end
[param,fold] = proc_argv(params); % specify each parameter
if fold <= 1
error(‘The number of folds must be greater than 1‘);
else
[deci,label_y] = get_cv_deci(y,x,param,fold); % get the value of decision and label after cross-calidation
auc = roc_curve(deci,label_y); % plot ROC curve
end
end
end
function [resu,fold] = proc_argv(params)
resu=params;
fold=5;
if ~isempty(params) && ~isempty(regexp(params,‘-v‘))
[fold_val,fold_start,fold_end] = regexp(params,‘-v\s+\d+‘,‘match‘,‘start‘,‘end‘);
if ~isempty(fold_val)
[temp1,fold] = strread([fold_val{:}],‘%s %u‘);
resu([fold_start:fold_end]) = [];
else
error(‘Number of CV folds must be specified by "-v cv_fold"‘);
end
end
end
function [deci,label_y] = get_cv_deci(prob_y,prob_x,param,nr_fold)
l=length(prob_y);
deci = ones(l,1);
label_y = ones(l,1);
rand_ind = randperm(l);
for i=1:nr_fold % Cross training : folding
test_ind=rand_ind([floor((i-1)*l/nr_fold)+1:floor(i*l/nr_fold)]‘);
train_ind = [1:l]‘;
train_ind(test_ind) = [];
model = svmtrain(prob_y(train_ind),prob_x(train_ind,:),param);
[predict_label,mse,subdeci] = svmpredict(prob_y(test_ind),prob_x(test_ind,:),model);
deci(test_ind) = subdeci.*model.Label(1);
label_y(test_ind) = prob_y(test_ind);
end
end
function auc = roc_curve(deci,label_y) %%deci=wx+b, label_y, true label
[val,ind] = sort(deci,‘descend‘);
roc_y = label_y(ind);
stack_x = cumsum(roc_y == -1)/sum(roc_y == -1);
stack_y = cumsum(roc_y == 1)/sum(roc_y == 1);
auc = sum((stack_x(2:length(roc_y),1)-stack_x(1:length(roc_y)-1,1)).*stack_y(2:length(roc_y),1))
%Comment the above lines if using perfcurve of statistics toolbox
%[stack_x,stack_y,thre,auc]=perfcurve(label_y,deci,1);
plot(stack_x,stack_y);
xlabel(‘False Positive Rate‘);
ylabel(‘True Positive Rate‘);
title([‘ROC curve of (AUC = ‘ num2str(auc) ‘ )‘]);
end
调用:
[y,x] = libsvmread(‘heart_scale.txt‘);
model = svmtrain(y,x);
plotroc(y,x,model);
标签:
原文地址:http://www.cnblogs.com/huadongw/p/5495004.html