标签:
resource: Evolutionary computing, A.E.Eiben
Evolution strategies(ES) is another member of the evolutionary algorithm family.
ES technical summary tableau
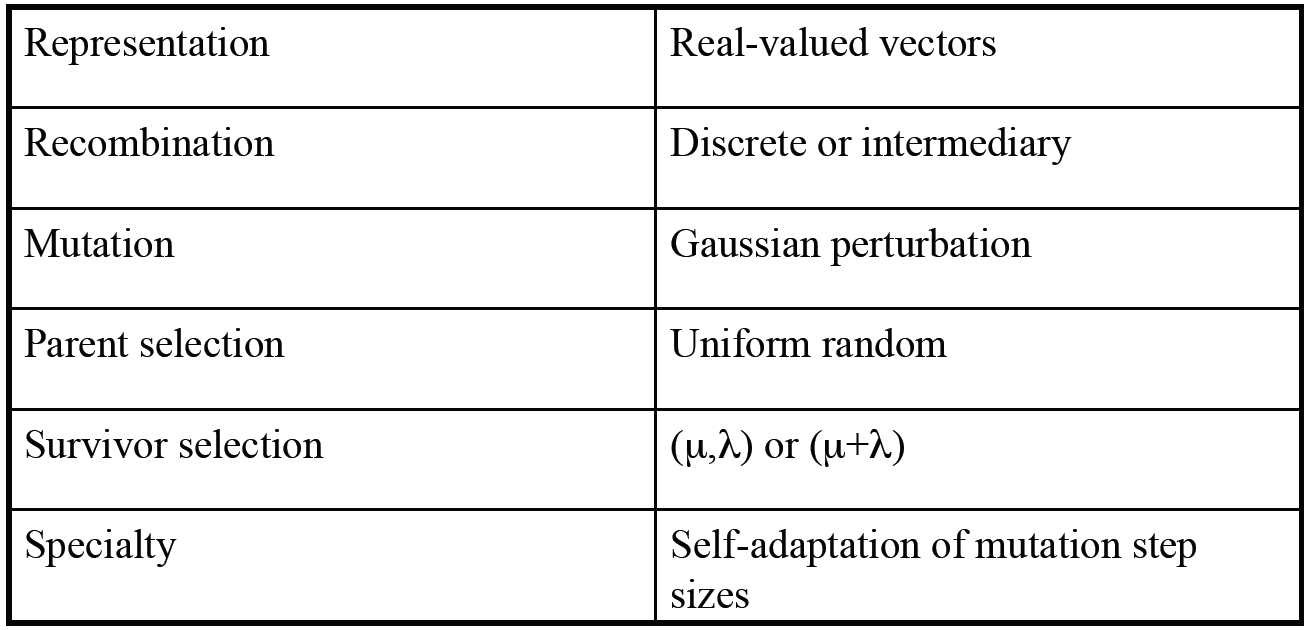
minimimise f : Rn -> R
“two-membered ES” using
------------------------------------------------------------
Set t = 0
Create initial point xt = 〈 x1t ,…,xnt 〉
REPEAT UNTIL (TERMIN.COND satisfied) DO
Draw zi from a normal distr. for all i = 1,…,n
yit = xit + zi
IF f(xt) < f(yt) THEN xt+1 = xt
ELSE xt+1 = yt
Set t = t+1
OD
------------------------------------------------------------
As is shown on the pseudocode above, given a current solution xt in the form of a vector of length n, a new candidate xt+1 is created by adding a random number zi for i =1,...,n to each of the n components.
( let‘s talk about the random number Zi )
The random number Zi:
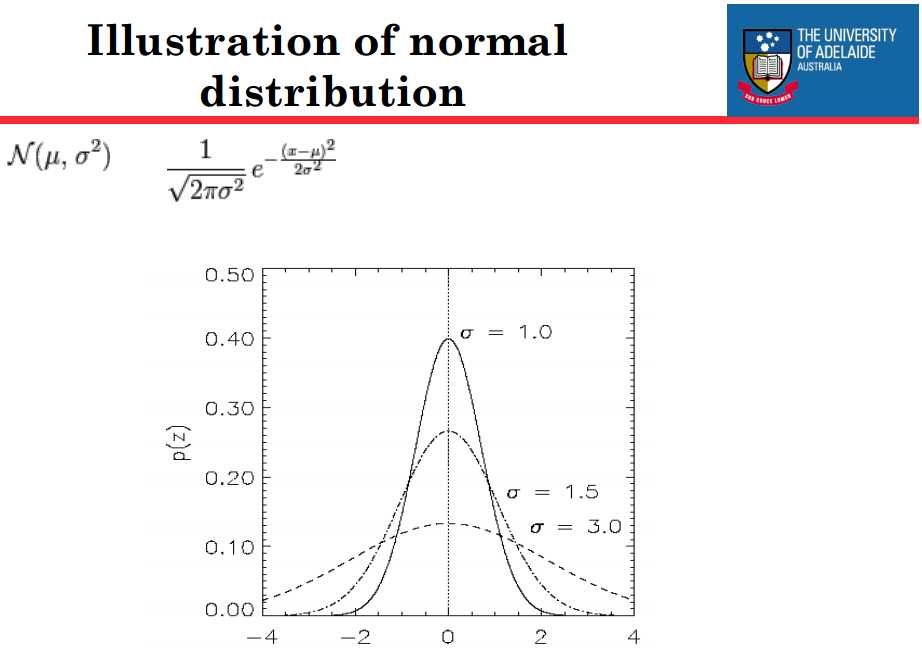
A Gaussian, or normal, distribution is used with zero mean and standard deviation σ for drawing the random numbers.
This distribution is symmetric about zero and has the feature that the probability of drawing a random number with any given magnitude is a rapidly decreasing function of the standard deviation σ. (more information about Gaussian distribution)
( let‘s talk about the random number σ)
Thus the σ value is a parameter of the algorithm that determines the extent to which given values xi are perturbed by the mutation operator.
For this reason σ is often called the mutation step size.
Theoretical studies motivated an on-line adjustment of step sizes by the famous 1/5 success rule. This rule states that the ratio of successful mutaions (those in which the child is fitter than the parent) to all mutations should be 1/5.
The rule is executed at periodic intervals.
For instance, after k iterations each σ is reset by
Where ps is the relative frequency of successful mutations measured over a number of trials, and the parameter c is in the range [0.817,1]
As is apparent, using this mechanism the step sizes change based on feedback from the search process.
This example illuminiates some essential characteristics of evolution strategies:
Chromosomes consist of three parts:
Full size: 〈 x1,…,xn, σ1,…,σn ,α1,…, αk 〉,where k = n(n-1)/2 (no. of i,j pairs) ---This is the general form of individuals in ES
The σ values represent the mutation step sizes, and their number nσ is usually either 1 or n. For any easonable self-adaptation mechanism at least one σ must be present.
The α values, which represent interactions between the step sizes used for different variables, are not always used.
Changing value by adding random noise drawn from normal distribution
The mutation operator in ES is based on a normal (Gaussian) distribution requiring two parameters: the mean ξ and the standard deviation σ.
Mutations then are realised by adding some Δxi to each xi, where the Δxi values are randomly drawn using the given Gaussian N(ξ,σ), with the corresponding probability density function.
xi‘ = xi + N(0,σ)
xi‘ can be seen as a new xi.
N(0,σ) here denotes a random number drawn from a Gaussian distribution with zero mean and standard deviation σ.
In the simplest case we would have one step size that applied to all the components xi and candidate solutions of the form <x1, ..., xn, σ>.
Mutations are then realised by replacing <x1, ..., xn, σ> by <x1‘, ..., xn‘, σ‘>, where σ‘ is the mutated value of σ and xi‘ = xi + N(0,σ)
The mutation step sizes are not set by the user; rather the σ is coevolving with the solutions.
In order to achieve this behaviour:
The rationale behind this is that a new individual <x‘, σ‘> is effectively evaluated twice:
This happens indirectly: a given step size (σ) evaluates favourably if the offspring generated by using it prove viable (in the first sense).
To sum up, an individual <x‘, σ‘> represents both a good x‘ that survived selection and a good σ‘ that proved successful in generating this good x‘ from x.
In the case of uncorrelated mutation with one step size, the same distribution is used to mutate each xi, therefore we only have one strategy parameter σ in each individual.
This σ is mutated each time step by multiplying it by a term eΓ, with Γ a random variable drawn each time from a normal distribution with mean 0 and standard deviation τ.
Since N(0,τ) = τ•N(0,1), the mutation mechanism is thus specified by the following formulas:
Furthermore, since standard deviations very close to zero are unwanted(they will have on average a negligible effect), the following boundary rule is used to force step sizes to be no smaller than a threshold:
Tips:
The proportionality constant τ is an external parameter to be set by the user.
It is usually inversely proportional to the square root of the problem size:
The parameter τ can be interpreted as a kind of learning rate, as in neural networks.
Evolutionary Computing: 5. Evolutionary Strategies
标签:
原文地址:http://www.cnblogs.com/adelaide/p/5738878.html