标签:算法实现 结构 batch 出发点 gre ilo 指定 pytho 距离
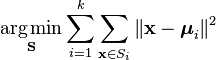
import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.clustering.KMeans; import org.apache.spark.mllib.clustering.KMeansModel; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; import org.apache.spark.SparkConf; public class KMeansExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("K-means Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse data String path = "data/mllib/kmeans_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); parsedData.cache(); // Cluster the data into two classes using KMeans int numClusters = 2; int numIterations = 20; KMeansModel clusters = KMeans.train(parsedData.rdd(), numClusters, numIterations); // Evaluate clustering by computing Within Set Sum of Squared Errors double WSSSE = clusters.computeCost(parsedData.rdd()); System.out.println("Within Set Sum of Squared Errors = " + WSSSE); // Save and load model clusters.save(sc.sc(), "myModelPath"); KMeansModel sameModel = KMeansModel.load(sc.sc(), "myModelPath"); } }
import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.clustering.GaussianMixture; import org.apache.spark.mllib.clustering.GaussianMixtureModel; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; import org.apache.spark.SparkConf; public class GaussianMixtureExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("GaussianMixture Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse data String path = "data/mllib/gmm_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.trim().split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); parsedData.cache(); // Cluster the data into two classes using GaussianMixture GaussianMixtureModel gmm = new GaussianMixture().setK(2).run(parsedData.rdd()); // Save and load GaussianMixtureModel gmm.save(sc.sc(), "myGMMModel"); GaussianMixtureModel sameModel = GaussianMixtureModel.load(sc.sc(), "myGMMModel"); // Output the parameters of the mixture model for(int j=0; j<gmm.k(); j++) { System.out.printf("weight=%f\nmu=%s\nsigma=\n%s\n", gmm.weights()[j], gmm.gaussians()[j].mu(), gmm.gaussians()[j].sigma()); } } }
import scala.Tuple2; import scala.Tuple3; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.clustering.PowerIterationClustering; import org.apache.spark.mllib.clustering.PowerIterationClusteringModel; // Load and parse the data JavaRDD<String> data = sc.textFile("data/mllib/pic_data.txt"); JavaRDD<Tuple3<Long, Long, Double>> similarities = data.map( new Function<String, Tuple3<Long, Long, Double>>() { public Tuple3<Long, Long, Double> call(String line) { String[] parts = line.split(" "); return new Tuple3<>(new Long(parts[0]), new Long(parts[1]), new Double(parts[2])); } } ); // Cluster the data into two classes using PowerIterationClustering PowerIterationClustering pic = new PowerIterationClustering() .setK(2) .setMaxIterations(10); PowerIterationClusteringModel model = pic.run(similarities); for (PowerIterationClustering.Assignment a: model.assignments().toJavaRDD().collect()) { System.out.println(a.id() + " -> " + a.cluster()); } // Save and load model model.save(sc.sc(), "myModelPath"); PowerIterationClusteringModel sameModel = PowerIterationClusteringModel.load(sc.sc(), "myModelPath");
maxIterations:EM迭代的最大次数topTopicsPerDocument:训练语料库中每个文档的顶级主题及其权重topDocumentsPerTopic: 每个主题的顶级文档和文档中主题的相应权重。logPrior:对数概率的估计问题,给出了文档的主题分布的参数各支流和topicconcentrationtopicConcentration:只有对称的先验知识的支持。值必须为= 0 = 0。在默认值为1的结果(1 K)(1 K)。topicConcentration:只有对称的先验知识的支持。值必须为= 0 = 0。在默认值为1的结果(1 K)(1 K)。maxIterations: 提交minibatches最大数。miniBatchFraction: 语料库样本和用于在每一次迭代的部分optimizeDocConcentration:如果设置为true,执行各支流的超参数的最大似然估计(又名α)在每个minibatch和套在返回的localldamodel优化各支tau0 and kappa: 用于学习率的衰减,这是由(τ0 +(0 +τITER)?κITER)?κ哪里iteriter是当前迭代次.logLikelihood(documents): 计算给定的给定推断主题的文档的下限.。logPerplexity(documents): 计算给定的推断的主题提供的文件的困惑的上限。import scala.Tuple2; import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.clustering.DistributedLDAModel; import org.apache.spark.mllib.clustering.LDA; import org.apache.spark.mllib.linalg.Matrix; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; import org.apache.spark.SparkConf; public class JavaLDAExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("LDA Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse the data String path = "data/mllib/sample_lda_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.trim().split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); // Index documents with unique IDs JavaPairRDD<Long, Vector> corpus = JavaPairRDD.fromJavaRDD(parsedData.zipWithIndex().map( new Function<Tuple2<Vector, Long>, Tuple2<Long, Vector>>() { public Tuple2<Long, Vector> call(Tuple2<Vector, Long> doc_id) { return doc_id.swap(); } } )); corpus.cache(); // Cluster the documents into three topics using LDA DistributedLDAModel ldaModel = new LDA().setK(3).run(corpus); // Output topics. Each is a distribution over words (matching word count vectors) System.out.println("Learned topics (as distributions over vocab of " + ldaModel.vocabSize() + " words):"); Matrix topics = ldaModel.topicsMatrix(); for (int topic = 0; topic < 3; topic++) { System.out.print("Topic " + topic + ":"); for (int word = 0; word < ldaModel.vocabSize(); word++) { System.out.print(" " + topics.apply(word, topic)); } System.out.println(); } ldaModel.save(sc.sc(), "myLDAModel"); DistributedLDAModel sameModel = DistributedLDAModel.load(sc.sc(), "myLDAModel"); }}
import com.google.common.collect.Lists; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.mllib.clustering.BisectingKMeans; import org.apache.spark.mllib.clustering.BisectingKMeansModel; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; ArrayList<Vector> localData = Lists.newArrayList( Vectors.dense(0.1, 0.1), Vectors.dense(0.3, 0.3), Vectors.dense(10.1, 10.1), Vectors.dense(10.3, 10.3), Vectors.dense(20.1, 20.1), Vectors.dense(20.3, 20.3), Vectors.dense(30.1, 30.1), Vectors.dense(30.3, 30.3));JavaRDD<Vector> data = sc.parallelize(localData, 2); BisectingKMeans bkm = new BisectingKMeans() .setK(4);BisectingKMeansModel model = bkm.run(data); System.out.println("Compute Cost: " + model.computeCost(data));for (Vector center: model.clusterCenters()) { System.out.println("");}Vector[] clusterCenters = model.clusterCenters();for (int i = 0; i < clusterCenters.length; i++) { Vector clusterCenter = clusterCenters[i]; System.out.println("Cluster Center " + i + ": " + clusterCenter); }

标签:算法实现 结构 batch 出发点 gre ilo 指定 pytho 距离
原文地址:http://www.cnblogs.com/yuguoshuo/p/6265762.html