%DT:DT实现根据乳腺肿瘤特征向量高精度预测肿瘤的是恶性还是良性
load data.mat
a = randperm(569);
Train = data(a(1:500),:);
Test = data(a(501:end),:);
P_train = Train(:,3:end);
T_train = Train(:,2);
P_test = Test(:,3:end);
T_test = Test(:,2);
ctree = ClassificationTree.fit(P_train,T_train);
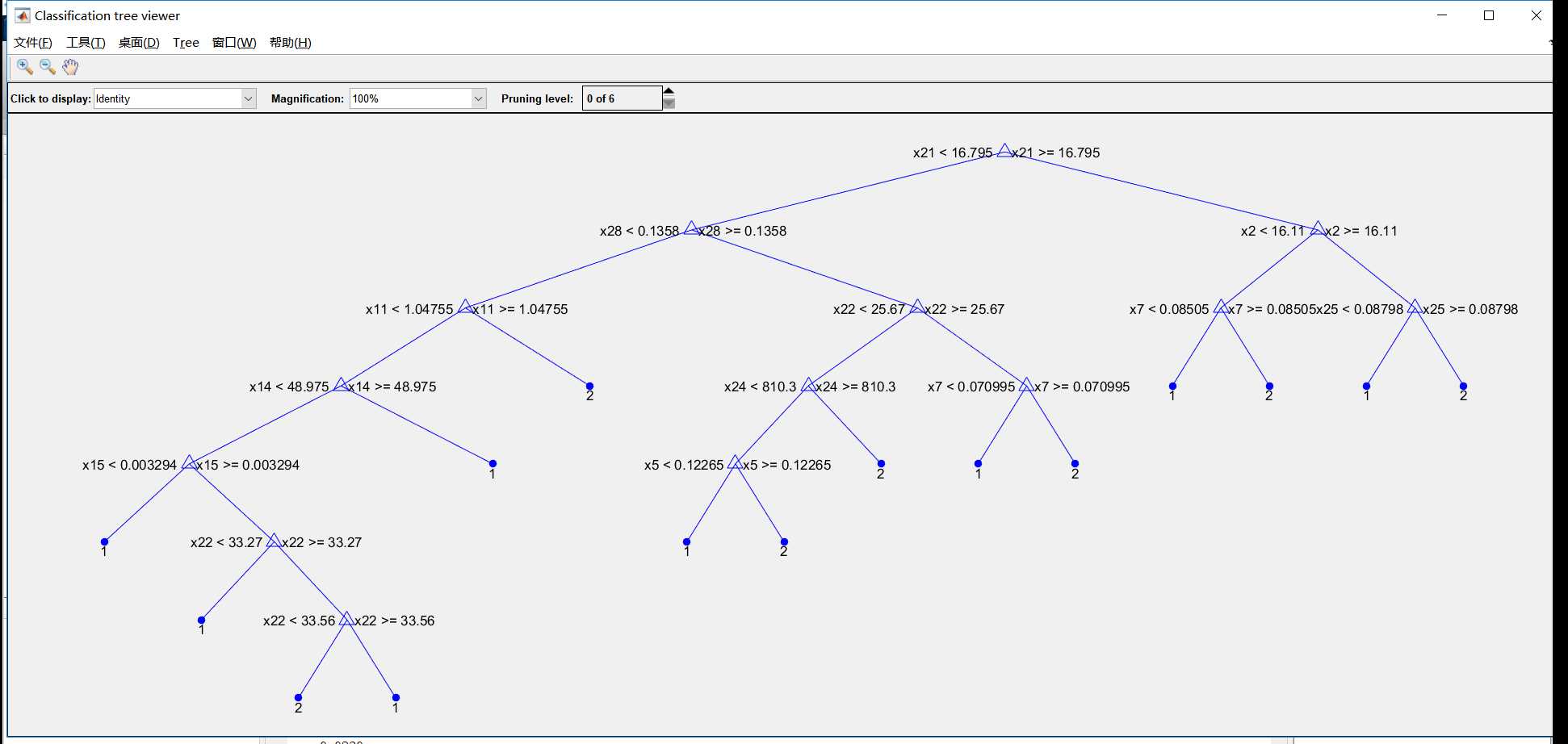
view(ctree);
view(ctree,‘mode‘,‘graph‘);
T_sim = predict(ctree,P_test);
count_B = length(find(T_train == 1));
count_M = length(find(T_train == 2));
rate_B = count_B / 500;
rate_M = count_M / 500;
total_B = length(find(data(:,2) == 1));
total_M = length(find(data(:,2) == 2));
number_B = length(find(T_test == 1));
number_M = length(find(T_test == 2));
number_B_sim = length(find(T_sim == 1 & T_test == 1));
number_M_sim = length(find(T_sim == 2 & T_test == 2));
disp([‘病例总数:‘ num2str(569)...
‘ 良性:‘ num2str(total_B)...
‘ 恶性:‘ num2str(total_M)]);
disp([‘训练集病例总数:‘ num2str(500)...
‘ 良性:‘ num2str(count_B)...
‘ 恶性:‘ num2str(count_M)]);
disp([‘测试集病例总数:‘ num2str(69)...
‘ 良性:‘ num2str(number_B)...
‘ 恶性:‘ num2str(number_M)]);
disp([‘良性乳腺肿瘤确诊:‘ num2str(number_B_sim)...
‘ 误诊:‘ num2str(number_B - number_B_sim)...
‘ 确诊率p1=‘ num2str(number_B_sim/number_B*100) ‘%‘]);
disp([‘恶性乳腺肿瘤确诊:‘ num2str(number_M_sim)...
‘ 误诊:‘ num2str(number_M - number_M_sim)...
‘ 确诊率p2=‘ num2str(number_M_sim/number_M*100) ‘%‘]);
disp([‘乳腺肿瘤整体预测准确率:‘ num2str((number_M_sim/number_M*100+number_B_sim/number_B*100)/2) ‘%‘]);
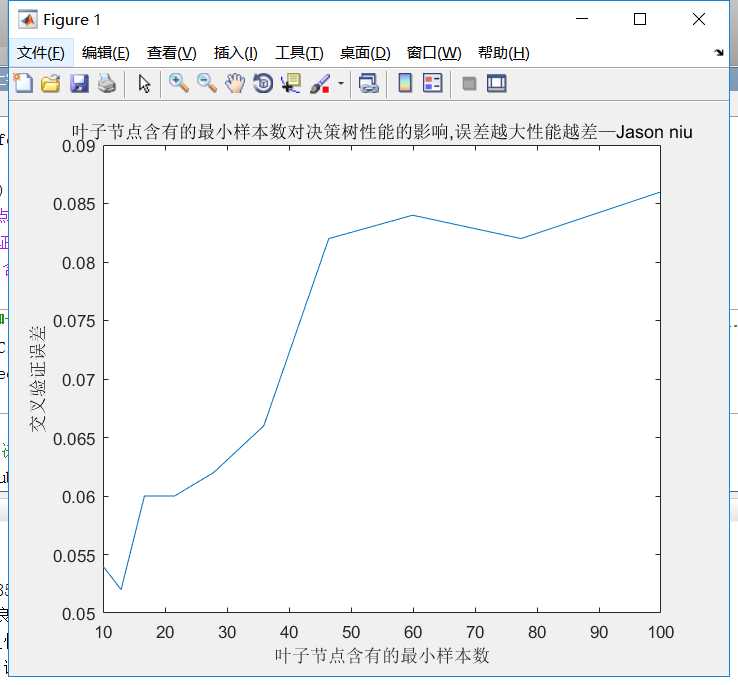
leafs = logspace(1,2,10);
N = numel(leafs);
err = zeros(N,1);
for n = 1:N
t = ClassificationTree.fit(P_train,T_train,‘crossval‘,‘on‘,‘minleaf‘,leafs(n));
err(n) = kfoldLoss(t);
end
plot(leafs,err);
xlabel(‘叶子节点含有的最小样本数‘);
ylabel(‘交叉验证误差‘);
title(‘叶子节点含有的最小样本数对决策树性能的影响,误差越大性能越差—Jason niu‘)
OptimalTree = ClassificationTree.fit(P_train,T_train,‘minleaf‘,13);
view(OptimalTree,‘mode‘,‘graph‘)
resubOpt = resubLoss(OptimalTree)
lossOpt = kfoldLoss(crossval(OptimalTree))
resubDefault = resubLoss(ctree)
lossDefault = kfoldLoss(crossval(ctree))
[~,~,~,bestlevel] = cvLoss(ctree,‘subtrees‘,‘all‘,‘treesize‘,‘min‘)
cptree = prune(ctree,‘Level‘,bestlevel);
view(cptree,‘mode‘,‘graph‘)
resubPrune = resubLoss(cptree)
lossPrune = kfoldLoss(crossval(cptree))