标签:cut erro sse back NPU specified use ace enc

import numpy path = r‘F:\数据分析专用\数据分析与机器学习\world_alcohol.txt‘ world_alchol = numpy.genfromtxt(path, delimiter=",", dtype=str) print(type(world_alchol)) print(world_alchol) print(help(numpy.genfromtxt))

<class ‘numpy.ndarray‘> [[‘Year‘ ‘WHO region‘ ‘Country‘ ‘Beverage Types‘ ‘Display Value‘] [‘1986‘ ‘Western Pacific‘ ‘Viet Nam‘ ‘Wine‘ ‘0‘] [‘1986‘ ‘Americas‘ ‘Uruguay‘ ‘Other‘ ‘0.5‘] ... [‘1987‘ ‘Africa‘ ‘Malawi‘ ‘Other‘ ‘0.75‘] [‘1989‘ ‘Americas‘ ‘Bahamas‘ ‘Wine‘ ‘1.5‘] [‘1985‘ ‘Africa‘ ‘Malawi‘ ‘Spirits‘ ‘0.31‘]] Help on function genfromtxt in module numpy.lib.npyio: genfromtxt(fname, dtype=<class ‘float‘>, comments=‘#‘, delimiter=None, skip_header=0, skip_footer=0, converters=None, missing_values=None, filling_values=None, usecols=None, names=None, excludelist=None, deletechars=None, replace_space=‘_‘, autostrip=False, case_sensitive=True, defaultfmt=‘f%i‘, unpack=None, usemask=False, loose=True, invalid_raise=True, max_rows=None, encoding=‘bytes‘) Load data from a text file, with missing values handled as specified. Each line past the first `skip_header` lines is split at the `delimiter` character, and characters following the `comments` character are discarded. Parameters ---------- fname : file, str, pathlib.Path, list of str, generator File, filename, list, or generator to read. If the filename extension is `.gz` or `.bz2`, the file is first decompressed. Note that generators must return byte strings in Python 3k. The strings in a list or produced by a generator are treated as lines. dtype : dtype, optional Data type of the resulting array. If None, the dtypes will be determined by the contents of each column, individually. comments : str, optional The character used to indicate the start of a comment. All the characters occurring on a line after a comment are discarded delimiter : str, int, or sequence, optional The string used to separate values. By default, any consecutive whitespaces act as delimiter. An integer or sequence of integers can also be provided as width(s) of each field. skiprows : int, optional `skiprows` was removed in numpy 1.10. Please use `skip_header` instead. skip_header : int, optional The number of lines to skip at the beginning of the file. skip_footer : int, optional The number of lines to skip at the end of the file. converters : variable, optional The set of functions that convert the data of a column to a value. The converters can also be used to provide a default value for missing data: ``converters = {3: lambda s: float(s or 0)}``. missing : variable, optional `missing` was removed in numpy 1.10. Please use `missing_values` instead. missing_values : variable, optional The set of strings corresponding to missing data. filling_values : variable, optional The set of values to be used as default when the data are missing. usecols : sequence, optional Which columns to read, with 0 being the first. For example, ``usecols = (1, 4, 5)`` will extract the 2nd, 5th and 6th columns. names : {None, True, str, sequence}, optional If `names` is True, the field names are read from the first line after the first `skip_header` lines. This line can optionally be proceeded by a comment delimeter. If `names` is a sequence or a single-string of comma-separated names, the names will be used to define the field names in a structured dtype. If `names` is None, the names of the dtype fields will be used, if any. excludelist : sequence, optional A list of names to exclude. This list is appended to the default list [‘return‘,‘file‘,‘print‘]. Excluded names are appended an underscore: for example, `file` would become `file_`. deletechars : str, optional A string combining invalid characters that must be deleted from the names. defaultfmt : str, optional A format used to define default field names, such as "f%i" or "f_%02i". autostrip : bool, optional Whether to automatically strip white spaces from the variables. replace_space : char, optional Character(s) used in replacement of white spaces in the variables names. By default, use a ‘_‘. case_sensitive : {True, False, ‘upper‘, ‘lower‘}, optional If True, field names are case sensitive. If False or ‘upper‘, field names are converted to upper case. If ‘lower‘, field names are converted to lower case. unpack : bool, optional If True, the returned array is transposed, so that arguments may be unpacked using ``x, y, z = loadtxt(...)`` usemask : bool, optional If True, return a masked array. If False, return a regular array. loose : bool, optional If True, do not raise errors for invalid values. invalid_raise : bool, optional If True, an exception is raised if an inconsistency is detected in the number of columns. If False, a warning is emitted and the offending lines are skipped. max_rows : int, optional The maximum number of rows to read. Must not be used with skip_footer at the same time. If given, the value must be at least 1. Default is to read the entire file. .. versionadded:: 1.10.0 encoding : str, optional Encoding used to decode the inputfile. Does not apply when `fname` is a file object. The special value ‘bytes‘ enables backward compatibility workarounds that ensure that you receive byte arrays when possible and passes latin1 encoded strings to converters. Override this value to receive unicode arrays and pass strings as input to converters. If set to None the system default is used. The default value is ‘bytes‘. .. versionadded:: 1.14.0 Returns ------- out : ndarray Data read from the text file. If `usemask` is True, this is a masked array. See Also -------- numpy.loadtxt : equivalent function when no data is missing. Notes ----- * When spaces are used as delimiters, or when no delimiter has been given as input, there should not be any missing data between two fields. * When the variables are named (either by a flexible dtype or with `names`, there must not be any header in the file (else a ValueError exception is raised). * Individual values are not stripped of spaces by default. When using a custom converter, make sure the function does remove spaces. References ---------- .. [1] NumPy User Guide, section `I/O with NumPy <http://docs.scipy.org/doc/numpy/user/basics.io.genfromtxt.html>`_. Examples --------- >>> from io import StringIO >>> import numpy as np Comma delimited file with mixed dtype >>> s = StringIO("1,1.3,abcde") >>> data = np.genfromtxt(s, dtype=[(‘myint‘,‘i8‘),(‘myfloat‘,‘f8‘), ... (‘mystring‘,‘S5‘)], delimiter=",") >>> data array((1, 1.3, ‘abcde‘), dtype=[(‘myint‘, ‘<i8‘), (‘myfloat‘, ‘<f8‘), (‘mystring‘, ‘|S5‘)]) Using dtype = None >>> s.seek(0) # needed for StringIO example only >>> data = np.genfromtxt(s, dtype=None, ... names = [‘myint‘,‘myfloat‘,‘mystring‘], delimiter=",") >>> data array((1, 1.3, ‘abcde‘), dtype=[(‘myint‘, ‘<i8‘), (‘myfloat‘, ‘<f8‘), (‘mystring‘, ‘|S5‘)]) Specifying dtype and names >>> s.seek(0) >>> data = np.genfromtxt(s, dtype="i8,f8,S5", ... names=[‘myint‘,‘myfloat‘,‘mystring‘], delimiter=",") >>> data array((1, 1.3, ‘abcde‘), dtype=[(‘myint‘, ‘<i8‘), (‘myfloat‘, ‘<f8‘), (‘mystring‘, ‘|S5‘)]) An example with fixed-width columns >>> s = StringIO("11.3abcde") >>> data = np.genfromtxt(s, dtype=None, names=[‘intvar‘,‘fltvar‘,‘strvar‘], ... delimiter=[1,3,5]) >>> data array((1, 1.3, ‘abcde‘), dtype=[(‘intvar‘, ‘<i8‘), (‘fltvar‘, ‘<f8‘), (‘strvar‘, ‘|S5‘)]) None
用array输入数组

vector = numpy.array([5, 10, 15, 20]) matrix = numpy.array([[5, 10, 15], [20, 25, 30], [35, 40, 45]]) print(vector) print(matrix)
输出结果

vector = numpy.array([1, 2, 3, 4]) print(vector.shape) matrix = numpy.array([[5, 10, 15], [20, 25, 30]]) print(matrix.shape)

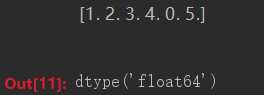
import numpy numbers = numpy.array([1, 2, 3, 4, 0, 5.0]) print(numbers) numbers.dtype

world_alchol = numpy.genfromtxt(path, delimiter=‘,‘, dtype=str, skip_header=1) print(world_alchol)

输出的是一个列表,那么读取的时候就可以根据切片读取出列表的值

uruguay_other_1986 = world_alchol[1, 4] third_country = world_alchol[2, 2] print(uruguay_other_1986) print(third_country)

标签:cut erro sse back NPU specified use ace enc
原文地址:https://www.cnblogs.com/pandaboy1123/p/9674208.html