标签:sum sam size cache save pre XSA .sh @param
1.算法简介
算法的原始论文 http://cs.nju.edu.cn/zhouzh/zhouzh.files/publication/icdm08b.pdf 。python的sklearn中已经实现了相关的api,对于单机的数据已经足够使用了,链接如下 http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.IsolationForest.html 。如果你想探究分布式下该算法怎么实现,下面细看。
按照惯例先讲一下算法的思想,对于已经了解的小伙伴来说,这段跳过。它的思路有点类似随机森林,并发训练N棵树,每棵树是没有关联的,且每棵树用到的样本和属性也是随机的,所不同的是,isolation forest (下面简称IF)是非监督的算法,通过构建二叉树,然后在构建好的树上,来预测样本的深度,如果深度太浅,则是疑似异常的样本。更加详细的论断和细节请查看论文,或者参考国内各大博客主写的个人感悟,我们把重点放在分布式实现上面。
2.分布式实现
在实现之前重点关注一点,IF算法并不需要所有的样本,甚至不能使用太多的样本,使用小样本的情况,算法效果更优。这一点在论文中有论断:
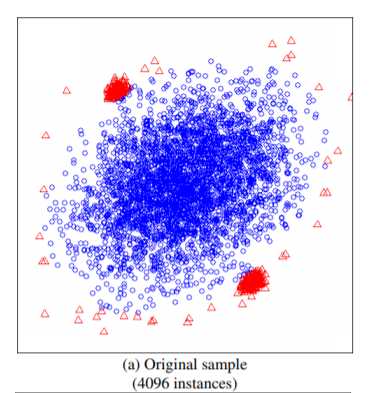
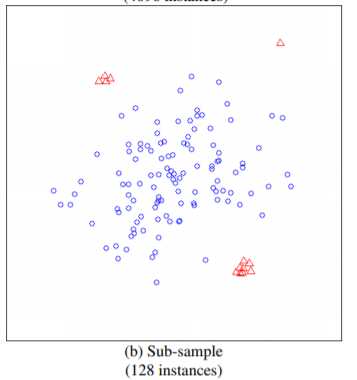
如上图所示,如果使用全部的样本作为训练集,则异常的样本,未必能识别出来,而在小样本下可以轻松识别。论文中比较了这两种方式,前者AUC达到0.67,而后者能达到0.91。
基于上面的论断,每棵树的样本大小不能太大,当然下面实现的方式既支持小样本又支持大样本,这个依赖于用户自己喜欢了
import java.util.concurrent.Executors import org.apache.spark.SparkContext import org.apache.spark.rdd.RDD import org.apache.spark.storage.StorageLevel import scala.concurrent.duration.Duration import scala.concurrent.{Await, ExecutionContext, Future} import scala.util.Random import org.apache.hadoop.fs._ sealed trait ITree case class ITreeBranch(left: ITree, right: ITree, split_column: Int, split_value: Double) extends ITree case class ITreeLeaf(size: Long) extends ITree /** @param trees trained trees * @param maxSamples The number of samples to train each base tree */ case class IForest(trees: Array[ITree], maxSamples: Int) { def predict(x: Array[Double]) = { if (trees.forall(_ == null)) { throw new Exception("Please train before predict!!") } else { val predictions = trees.map(s => pathLength(x, s, 0)).toList math.pow(2, -(predictions.sum / predictions.size) / cost(maxSamples)) } } @scala.annotation.tailrec final def pathLength(x: Array[Double], tree: ITree, path_length: Int): Double = { tree match { case ITreeLeaf(size) => path_length + cost(size) case ITreeBranch(left, right, split_column, split_value) => val sample_value = x(split_column) if (sample_value < split_value) pathLength(x, left, path_length + 1) else pathLength(x, right, path_length + 1) } } private def cost(num_items: Long): Double = if (num_items <= 1) 1.0 else 2.0 * (math.log(num_items - 1.0) + 0.577215664901532860606512090082402431) - (2.0 * (num_items - 1.0) / num_items) } object IForest { /** * @param numTrees The number of base tree in the ensemble * @param maxSamples The number of samples to train each base tree ,should be small!! should be small!! should be small!! * should be small!! should be small!! should be small!! * @param maxFeatures The fraction of features to train each base tree value in (0.0,1.0] * // * @param withReplacement whether sampling is done with replacement, do something in future * @param nJobs The number of jobs to run in parallel for fit ,do something in future */ def buildForest(data: RDD[Array[Double]], numTrees: Int = 100, maxSamples: Int = 256, maxFeatures: Double = 1.0, nJobs: Int = 10, distribute: Boolean = false) = { val sc = data.sparkContext val cacheData = if (sc.getRDDStorageInfo.filter(_.id == data.id).nonEmpty) data else data.persist(StorageLevel.MEMORY_AND_DISK) val dataCnt = data.count() println(s"AllSmaples =>${dataCnt}") val numFeatures = cacheData.take(1)(0).size checkData(cacheData, numFeatures) val sampleNumSamples = Math.min(maxSamples, dataCnt).toInt val sampleNumFeatures = (maxFeatures * numFeatures).toInt val maxDepth = Math.ceil((math.log(math.max(sampleNumSamples, 2)) / math.log(2))).toInt val sampleRatio = Math.min(sampleNumSamples * 1.0 / dataCnt * 2, 1.0) val trees = if (distribute) { implicit val xc = ExecutionContext.fromExecutorService(Executors.newFixedThreadPool(nJobs)) val tasks = (0 until numTrees).map { i => val sampleData = cacheData.sample(false, sampleRatio, System.currentTimeMillis()).zipWithIndex().filter(_._2 <= sampleNumSamples).map(_._1) parallizeGrow(sampleData, maxDepth, numFeatures, sampleNumFeatures) } val results = Await.result(Future.sequence(tasks), Duration.Inf) results.toArray } else (0 until numTrees).sliding(nJobs, nJobs).map { arr => sc.union( arr.map { i => cacheData.sample(false, sampleRatio, System.currentTimeMillis()).zipWithIndex().filter(_._2 <= sampleNumSamples) .map(_._1).repartition(1).mapPartitions { iter => val delta = iter.toArray val sampleFeatures = if (sampleNumFeatures < numFeatures) Random.shuffle((0 until numFeatures).toList).take(sampleNumFeatures) else (0 until numFeatures).toList Iterator(growTree(delta, maxDepth, sampleFeatures, 0)) } } ).collect() }.reduce(_ ++ _) new IForest(trees, maxSamples) } def saveModel(sc: SparkContext, iforest: IForest, path: String) = { val hdfs=FileSystem.get(sc.hadoopConfiguration) hdfs.delete(new Path(path), true) sc.parallelize(Seq(iforest), 1).saveAsObjectFile(path) } def loadModel(sc: SparkContext, path: String) = { sc.objectFile[IForest](path).collect()(0) } private def growTree(data: Array[Array[Double]], maxDepth: Int, sampleFeatures: Seq[Int], currentDepth: Int): ITree = { val numSamples = data.length if (currentDepth >= maxDepth || numSamples <= 1 || data.distinct.length == 1) { new ITreeLeaf(numSamples) } else { val splitColumn = sampleFeatures(Random.nextInt(sampleFeatures.length)) val columnValue = data.map(_.apply(splitColumn)) val colMin = columnValue.min val colMax = columnValue.max val splitValue = colMin + Random.nextDouble() * (colMax - colMin) val dataLeft = data.filter(_ (splitColumn) < splitValue) val dataRight = data.filter(_ (splitColumn) >= splitValue) new ITreeBranch(growTree(dataLeft, maxDepth, sampleFeatures, currentDepth + 1), growTree(dataRight, maxDepth, sampleFeatures, currentDepth + 1), splitColumn, splitValue) } } private def parallizeGrow(data: RDD[Array[Double]], maxDepth: Int, numFeatures: Int, sampleNumFeatures: Int)(implicit xc: ExecutionContext) = Future { val sampleFeatures = if (sampleNumFeatures < numFeatures) Random.shuffle((0 until numFeatures).toList).take(sampleNumFeatures) else (0 until numFeatures) growTree(data, maxDepth, sampleFeatures, 0) } private def growTree(data: RDD[Array[Double]], maxDepth: Int, sampleFeatures: Seq[Int], currentDepth: Int): ITree = { val sc = data.sparkContext val cacheData = if (sc.getRDDStorageInfo.filter(_.id == data.id).length > 0) data else data.persist(StorageLevel.MEMORY_AND_DISK) val numSamples = cacheData.count() val ret = if (currentDepth >= maxDepth || numSamples <= 1 || cacheData.distinct.count() == 1) { new ITreeLeaf(numSamples) } else { val splitColumn = sampleFeatures(Random.nextInt(sampleFeatures.length)) val columnValue = cacheData.map(_ (splitColumn)) val colMin = columnValue.min() val colMax = columnValue.max() val splitValue = colMin + Random.nextDouble() * (colMax - colMin) val dataLeft = cacheData.filter(_ (splitColumn) < splitValue) val dataRight = cacheData.filter(_ (splitColumn) >= splitValue) new ITreeBranch(growTree(dataLeft, maxDepth, sampleFeatures, currentDepth + 1), growTree(dataRight, maxDepth, sampleFeatures, currentDepth + 1), splitColumn, splitValue) } cacheData.unpersist() ret } private def checkData(data: RDD[Array[Double]], numFeatures: Int) = { assert(data.filter(arr => !(arr.length == numFeatures)).isEmpty(), "data must in equal column size") } }
代码说明:
3.总结
1.代码已经测试通过,直接mvn编译打包,运行环境为hadoop3.1.0和spark2.3,大家放心使用。
2.如有疑问欢迎指正,大家相互学习交流。
【异常检测】Isolation forest 的spark 分布式实现
标签:sum sam size cache save pre XSA .sh @param
原文地址:https://www.cnblogs.com/wzyj/p/9696357.html