标签:cto ges install file bit clu led 图片 efs
一句话:我们经常会面对非模式物种的GO或者KEGG富集与注释。
if("clusterProfiler" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("clusterProfiler")}
biocLite("colorspace")
suppressMessages(library(clusterProfiler))
ls("package:clusterProfiler")
library(AnnotationHub)
library(biomaRt)
library(enrichGO)
hub <- AnnotationHub::AnnotationHub() query(hub, "Apis cerana")
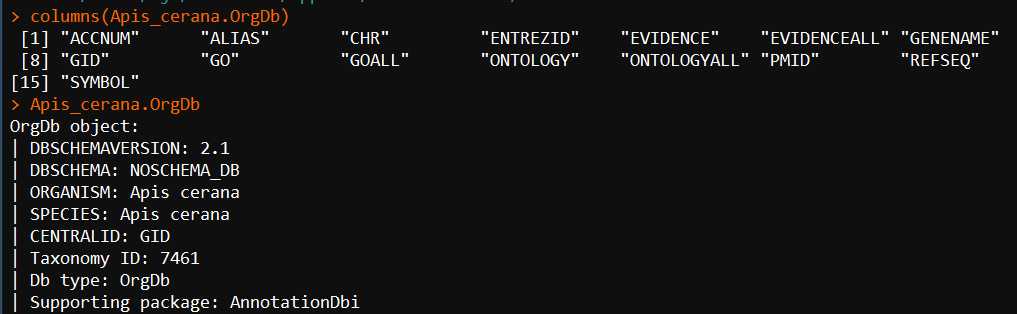
Apis_cerana.OrgDb <- hub[["AH62635"]] columns(Apis_cerana.OrgDb) Apis_cerana.OrgDb
keys(Apis_cerana.OrgDb)
head(keys(Apis_cerana.OrgDb,keytype = "SYMBOL")) # 查看注释信息的symbol
hainan_genelist =read.table(‘C:/Users/djx/Desktop/海南.txt‘,header=TRUE,sep=‘\t‘)
head(hainan_genelist)
colnames(hainan_genelist)
rownames(hainan_genelist)
bitr(keys(Apis_cerana.OrgDb)[15], ‘SYMBOL‘, c("ENTREZID","REFSEQ", "GO", "ONTOLOGY"), Apis_cerana.OrgDb)
hainan_geneid=as.character(hainan_genelist$Gene.ID)
hainan_geneid[1]
str(hainan_geneid)
rm(hainan_geneid)
sample_genes <- keys(Apis_cerana.OrgDb)[1:100]
hainan_enrich.go = enrichGO(sample_genes,
OrgDb = Apis_cerana.OrgDb,
keyType = ‘ENTREZID‘,
#ont= "BP",
pvalueCutoff = 0.05,
qvalueCutoff = 0.1,
readable = T)
标签:cto ges install file bit clu led 图片 efs
原文地址:https://www.cnblogs.com/djx571/p/10271874.html