标签:style blog http color io os ar for sp
核函数估计
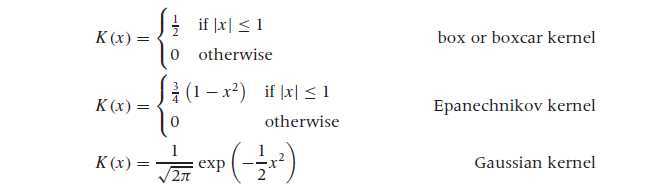
Demo I
import sys,re,os import numpy as np from scipy import stats import matplotlib.pylab as plt if __name__ == ‘__main__‘: # random data grade = [np.random.rand(100) * 100] fig = plt.figure() # KDE ax1 = fig.add_subplot(211) ind = np.arange(0.,100.,1) gkde = stats.kde.gaussian_kde(grade, bw_method = ‘scott‘) ax1.plot(ind, gkde(ind), label=‘Gods\‘ Grade‘, color="g") ax1.set_title(‘Kernel Density Estimation‘) ax1.legend() # hisogram ax2 = fig.add_subplot(212) ax2.hist(grade, 100, range = (0,100), normed = True) plt.show()
Demo II
scikit-learn中的demo
http://scikit-learn.org/stable/auto_examples/neighbors/plot_kde_1d.html
# -*- coding: utf-8 -*- """ Created on Wed Oct 22 20:38:13 2014 @author: dell """ # Author: Jake Vanderplas <jakevdp@cs.washington.edu> # import numpy as np import matplotlib.pyplot as plt from scipy.stats import norm from sklearn.neighbors import KernelDensity #---------------------------------------------------------------------- # Plot the progression of histograms to kernels np.random.seed(1) N = 20 X = np.concatenate((np.random.normal(0, 1, 0.3 * N), np.random.normal(5, 1, 0.7 * N)))[:, np.newaxis] X_plot = np.linspace(-5, 10, 1000)[:, np.newaxis] bins = np.linspace(-5, 10, 10) fig, ax = plt.subplots(2, 2, sharex=True, sharey=True) fig.subplots_adjust(hspace=0.05, wspace=0.05) # histogram 1 ax[0, 0].hist(X[:, 0], bins=bins, fc=‘#AAAAFF‘, normed=True) ax[0, 0].text(-3.5, 0.31, "Histogram") # histogram 2 ax[0, 1].hist(X[:, 0], bins=bins + 0.75, fc=‘#AAAAFF‘, normed=True) ax[0, 1].text(-3.5, 0.31, "Histogram, bins shifted") # tophat KDE kde = KernelDensity(kernel=‘tophat‘, bandwidth=0.75).fit(X) log_dens = kde.score_samples(X_plot) ax[1, 0].fill(X_plot[:, 0], np.exp(log_dens), fc=‘#AAAAFF‘) ax[1, 0].text(-3.5, 0.31, "Tophat Kernel Density") # Gaussian KDE kde = KernelDensity(kernel=‘gaussian‘, bandwidth=0.75).fit(X) log_dens = kde.score_samples(X_plot) ax[1, 1].fill(X_plot[:, 0], np.exp(log_dens), fc=‘#AAAAFF‘) ax[1, 1].text(-3.5, 0.31, "Gaussian Kernel Density") for axi in ax.ravel(): axi.plot(X[:, 0], np.zeros(X.shape[0]) - 0.01, ‘+k‘) axi.set_xlim(-4, 9) axi.set_ylim(-0.02, 0.34) for axi in ax[:, 0]: axi.set_ylabel(‘Normalized Density‘) for axi in ax[1, :]: axi.set_xlabel(‘x‘) #---------------------------------------------------------------------- # Plot all available kernels X_plot = np.linspace(-6, 6, 1000)[:, None] X_src = np.zeros((1, 1)) fig, ax = plt.subplots(2, 3, sharex=True, sharey=True) fig.subplots_adjust(left=0.05, right=0.95, hspace=0.05, wspace=0.05) def format_func(x, loc): if x == 0: return ‘0‘ elif x == 1: return ‘h‘ elif x == -1: return ‘-h‘ else: return ‘%ih‘ % x for i, kernel in enumerate([‘gaussian‘, ‘tophat‘, ‘epanechnikov‘, ‘exponential‘, ‘linear‘, ‘cosine‘]): axi = ax.ravel()[i] log_dens = KernelDensity(kernel=kernel).fit(X_src).score_samples(X_plot) axi.fill(X_plot[:, 0], np.exp(log_dens), ‘-k‘, fc=‘#AAAAFF‘) axi.text(-2.6, 0.95, kernel) axi.xaxis.set_major_formatter(plt.FuncFormatter(format_func)) axi.xaxis.set_major_locator(plt.MultipleLocator(1)) axi.yaxis.set_major_locator(plt.NullLocator()) axi.set_ylim(0, 1.05) axi.set_xlim(-2.9, 2.9) ax[0, 1].set_title(‘Available Kernels‘) #---------------------------------------------------------------------- # Plot a 1D density example N = 100 np.random.seed(1) X = np.concatenate((np.random.normal(0, 1, 0.3 * N), np.random.normal(5, 1, 0.7 * N)))[:, np.newaxis] X_plot = np.linspace(-5, 10, 1000)[:, np.newaxis] true_dens = (0.3 * norm(0, 1).pdf(X_plot[:, 0]) + 0.7 * norm(5, 1).pdf(X_plot[:, 0])) fig, ax = plt.subplots() ax.fill(X_plot[:, 0], true_dens, fc=‘black‘, alpha=0.2, label=‘input distribution‘) for kernel in [‘gaussian‘, ‘tophat‘, ‘epanechnikov‘]: kde = KernelDensity(kernel=kernel, bandwidth=0.5).fit(X) log_dens = kde.score_samples(X_plot) ax.plot(X_plot[:, 0], np.exp(log_dens), ‘-‘, label="kernel = ‘{0}‘".format(kernel)) ax.text(6, 0.38, "N={0} points".format(N)) ax.legend(loc=‘upper left‘) ax.plot(X[:, 0], -0.005 - 0.01 * np.random.random(X.shape[0]), ‘+k‘) ax.set_xlim(-4, 9) ax.set_ylim(-0.02, 0.4) plt.show()
PIC2, Kernel Density Estimates
标签:style blog http color io os ar for sp
原文地址:http://www.cnblogs.com/hluo/p/4044241.html