标签:lse data sed scene model number oda arm udf
首发于“生信补给站” ,https://mp.weixin.qq.com/s/l9Ci7wREQWpEV5dTvKuoHg,更多的R统计,绘图,生信,请移步??
msleep数据集,介绍了列的操作,盘一盘Tidyverse| 筛行选列之select,玩转列操作#载入R包
#install.packages("tidyverse")
library("tidyverse")
#查看内置数据集
head(msleep,2)
dim(msleep)
#[1] 83 11
本示例数据集很小,实际中数据量很大,可以根据使用filter()函数筛选出后续需要的“行”子集。
基于数值,根据 >, >=, <, <=, == 和 !=完成筛选
msleep %>% select(name, sleep_total) %>%
filter(sleep_total > 18) %>% head(2)
# A tibble: 2 x 2
name sleep_total
<chr> <dbl>
1 Big brown bat 19.7
2 Thick-tailed opposum 19.4
可以filter(sleep_total >= 16, sleep_total <= 18),也可以使用between函数
msleep %>% select(name, sleep_total) %>%
filter(between(sleep_total, 16, 18)) %>% head(2)
# A tibble: 2 x 2
name sleep_total
<chr> <dbl>
1 Owl monkey 17
2 Long-nosed armadillo 17.4
near()选择”中心值“,tol()设定范围,可以是值,可以是函数
msleep %>% select(name, sleep_total) %>%
filter(near(sleep_total, mean(sleep_total), tol = 3*sd(sleep_total))) %>% head(2)
# A tibble: 83 x 2
name sleep_total
<chr> <dbl>
1 Cheetah 12.1
2 Owl monkey 17
msleep %>% select(order, name, sleep_total) %>%
filter(order == "Didelphimorphia")
## # A tibble: 2 x 3
## order name sleep_total
## <chr> <chr> <dbl>
## 1 Didelphimorphia North American Opossum 18.0
## 2 Didelphimorphia Thick-tailed opposum 19.4
不等于方式: filter(order != "Rodentia")
%in%实现多个变量选择
msleep %>% select(order, name, sleep_total) %>%
filter(order %in% c("Didelphimorphia", "Diprotodontia"))
# A tibble: 4 x 3
order name sleep_total
<chr> <chr> <dbl>
1 Didelphimorphia North American Opossum 18
2 Didelphimorphia Thick-tailed opposum 19.4
3 Diprotodontia Phalanger 13.7
4 Diprotodontia Potoroo 11.1
注意区分,此处 !%in%无作用! 需要在order前面加“!”
msleep %>% select(order, name, sleep_total) %>%
filter(! order %in% c("Didelphimorphia", "Diprotodontia"))
# A tibble: 79 x 3
order name sleep_total
<chr> <chr> <dbl>
1 Carnivora Cheetah 12.1
2 Primates Owl monkey 17
两个主要函数:基础包的 grepl() 函数, stringr 包的 str_detect() 函数
msleep %>% select(name, sleep_total) %>%
filter(str_detect(tolower(name), pattern = "mouse"))
# A tibble: 5 x 2
name sleep_total
<chr> <dbl>
1 Vesper mouse 7
2 House mouse 12.5
注意:正则匹配时,R区分大小写!
filter(condition1, condition2) 两个条件都符合;
filter(condition1, !condition2) 条件1是TRUE,条件2是FALSE;
filter(condition1 | condition2) 两个条件符合其中一个即可;
filter(xor(condition1, condition2) 只有一个条件符合!两个都符合不可以。注意使用
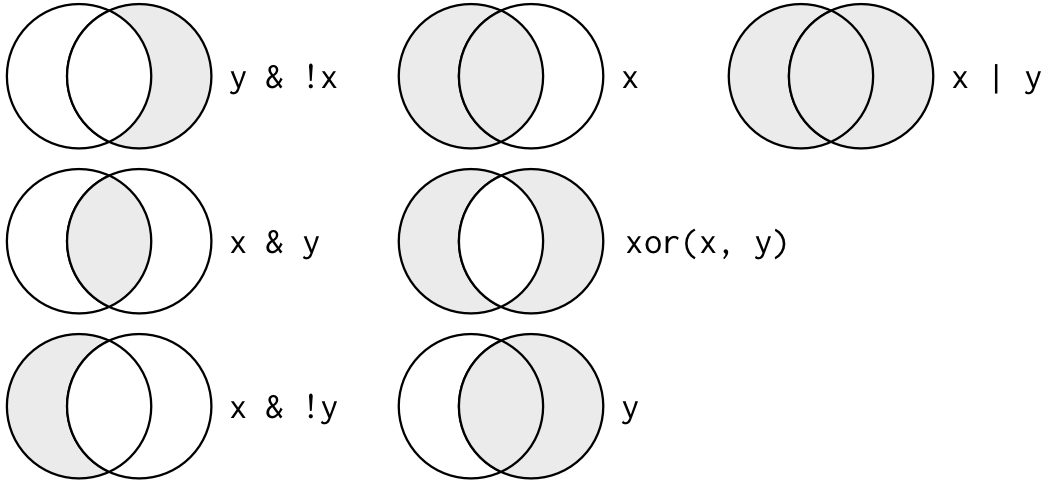
1)筛选 (bodywt大于100 ) 且 (sleep_total大于15 或者 order不为Carnivora )
msleep %>% select(name, order, sleep_total:bodywt) %>%
filter(bodywt > 100, (sleep_total > 15 | order != "Carnivora")) %>% head(2)
# A tibble: 2 x 8
name order sleep_total sleep_rem sleep_cycle awake brainwt bodywt
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Cow Artiodactyla 4 0.7 0.667 20 0.423 600
2 Asian elephant Proboscidea 3.9 NA NA 20.1 4.60 2547
额 ,注意逻辑!
注:多个条件可以用AND, OR ,NOT 组合
is.na内置完成 NA的筛选
#去除conservation是NA的所有行
msleep %>% select(name, conservation:sleep_cycle) %>%
filter(!is.na(conservation))
# A tibble: 54 x 5
name conservation sleep_total sleep_rem sleep_cycle
<chr> <chr> <dbl> <dbl> <dbl>
1 Cheetah lc 12.1 NA NA
2 Mountain beaver nt 14.4 2.4 NA
filter_all() 所有列参与筛选,根据筛选条件筛选
filter_if() 逻辑判断确定哪些列参与筛选,根据筛选条件筛选
filter_at() vars()函数定义哪些列参与筛选,根据筛选条件筛选
首先指定哪些列,然后确定筛选器的条件,多数情况下,需要.操作符引向待筛选的值。
filter_all()筛选所有行#筛选name:order, sleep_total, -vore中,任何一列包含“Ca”的所有行
msleep %>% select(name:order, sleep_total, -vore) %>%
filter_all(any_vars(str_detect(., pattern = "Ca")))
# A tibble: 16 x 4
name genus order sleep_total
<chr> <chr> <chr> <dbl>
1 Cheetah Acinonyx Carnivora 12.1
2 Northern fur seal Callorhinus Carnivora 8.7
注: any_vars() 类似OR,all_vars()则类似AND
#筛选name, sleep_total:bodywt, -awake列中,所有列均大于1的所有行
msleep %>% select(name, sleep_total:bodywt, -awake) %>%
filter_all(all_vars(. > 1))
# A tibble: 1 x 6
name sleep_total sleep_rem sleep_cycle brainwt bodywt
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Human 8 1.9 1.5 1.32 62
filter_if()筛选条件列msleep %>%
select(name:order, sleep_total:sleep_rem) %>%
filter_if(is.character, any_vars(is.na(.)))
# A tibble: 7 x 6
name genus vore order sleep_total sleep_rem
<chr> <chr> <chr> <chr> <dbl> <dbl>
1 Vesper mouse Calomys NA Rodentia 7 NA
2 Desert hedgehog Paraechinus NA Erinaceomorpha 10.3 2.7
注:同样可以使用 is.numeric, is.integer, is.double, is.logical, is.factor判别,日期数据的话,可以加载 lubridate 包,使用 is.POSIXt 或者 is.Date.
filter_at()筛选条件列优点:自定义待筛选的列,无需指定待筛选的列的类型
#筛选sleep_total, sleep_rem两个变量,所有值均大于5的行
msleep %>%
select(name, sleep_total:sleep_rem, brainwt:bodywt) %>%
filter_at(vars(sleep_total, sleep_rem), all_vars(.>5))
# A tibble: 2 x 5
name sleep_total sleep_rem brainwt bodywt
<chr> <dbl> <dbl> <dbl> <dbl>
1 Thick-tailed opposum 19.4 6.6 NA 0.37
2 Giant armadillo 18.1 6.1 0.081 60
当然也可以使用正则表达式
msleep %>%
select(name, sleep_total:sleep_rem, brainwt:bodywt) %>%
filter_at(vars(contains("sleep")), all_vars(.>5))
# A tibble: 2 x 5
name sleep_total sleep_rem brainwt bodywt
<chr> <dbl> <dbl> <dbl> <dbl>
1 Thick-tailed opposum 19.4 6.6 NA 0.37
2 Giant armadillo 18.1 6.1 0.081 60
好了,筛选行大概介绍这么多,应该能解决大部分筛选问题。
《R数据科学》
https://r4ds.had.co.nz/introduction.html
https://suzanbaert.netlify.com/2018/01/dplyr-tutorial-1/
◆ ◆ ◆ ◆ ◆
盘一盘Tidyverse| 只要你要只要我有-filter 筛选行
标签:lse data sed scene model number oda arm udf
原文地址:https://www.cnblogs.com/Mao1518202/p/12393984.html