标签:
Bioconductor的两个基础类:ExpressionSet类和SummarizedExpression类
ExpressionSet类和SummarizedExpression类是储存高通量数据的两个基础类。ExpressionSet主要用于基于array的研究,它的row是feature,而SummarizedExpression主要用于基于测序的研究,它的row是genomic ranges(GRanges)。
ExpressionSet和SummarizedExpression的操作
ExpressionSet类位于Biobase的包中,我们以“GSE9514”这个microarray的数据为例。加载GEOquery可以使用GSE的ID从GEO下载对应数据。
library(Biobase)
library(GEOquery)
geoq <- getGEO("GSE9514")
这里geoq是只有一个元素的list。
names(geoq) #[1] "GSE9514_series_matrix.txt.gz" e <- geoq[[1]] e # ExpressionSet (storageMode: lockedEnvironment) # assayData: 9335 features, 8 samples # element names: exprs # protocolData: none # phenoData # sampleNames: GSM241146 GSM241147 ... GSM241153 (8 total) # varLabels: title geo_accession ... data_row_count (31 total) # varMetadata: labelDescription # featureData # featureNames: 10000_at 10001_at ... AFFX-YFL039CM_at (9335 total) # fvarLabels: ID ORF ... Gene Ontology Molecular Function (17 total) # fvarMetadata: Column Description labelDescription # experimentData: use ‘experimentData(object)‘ # Annotation: GPL90
e就是我们需要的ExpressionSet。通过expr函数可以提取表达值矩阵,同时可使用feature或gene的名称对矩阵索引。
exprs(e)[1:3,1:3] # GSM241146 GSM241147 GSM241148 # 10000_at 15.33081 9.459372 7.984865 # 10001_at 283.47190 300.729460 270.016080 # 10002_i_at 2569.45360 2382.814700 2711.814500 dim(exprs(e)) # [1] 9335 8
pData和fData函数可以提取表型(feature)和基因信息,对应于表达矩阵的column和row,这两个对象的操作类似于DataFrame。
pData(e)[1:3,1:6] # title geo_accession # GSM241146 hem1 strain grown in YPD with 250 uM ALA (08-15-06_Philpott_YG_S98_1) GSM241146 # GSM241147 WT strain grown in YPD under Hypoxia (08-15-06_Philpott_YG_S98_10) GSM241147 # GSM241148 WT strain grown in YPD under Hypoxia (08-15-06_Philpott_YG_S98_11) GSM241148 # status submission_date last_update_date type # GSM241146 Public on Nov 06 2007 Nov 02 2007 Aug 14 2011 RNA # GSM241147 Public on Nov 06 2007 Nov 02 2007 Aug 14 2011 RNA # GSM241148 Public on Nov 06 2007 Nov 02 2007 Aug 14 2011 RNA dim(pData(e)) # [1] 8 31 names(pData(e)) # [1] "title" "geo_accession" "status" # [4] "submission_date" "last_update_date" "type" # [7] "channel_count" "source_name_ch1" "organism_ch1" # [10] "characteristics_ch1" "molecule_ch1" "extract_protocol_ch1" # [13] "label_ch1" "label_protocol_ch1" "taxid_ch1" # [16] "hyb_protocol" "scan_protocol" "description" # [19] "data_processing" "platform_id" "contact_name" # [22] "contact_email" "contact_department" "contact_institute" # [25] "contact_address" "contact_city" "contact_state" # [28] "contact_zip/postal_code" "contact_country" "supplementary_file" # [31] "data_row_count" pData(e)$characteristics_ch1 # V2 V3 V4 V5 # BY4742 hem1 delta strain BY4742 BY4742 BY4742 hem1 delta strain # V6 V7 V8 V9 # BY4742 hem1 delta strain BY4742 hem1 delta strain BY4742 strain BY4742 strain # Levels: BY4742 BY4742 hem1 delta strain BY4742 strain fData(e)[1:3,1:3] # ID ORF SPOT_ID # 10000_at 10000_at YLR331C <NA> # 10001_at 10001_at YLR332W <NA> # 10002_i_at 10002_i_at YLR333C <NA> dim(fData(e)) # [1] 9335 17 names(fData(e)) # [1] "ID" "ORF" # [3] "SPOT_ID" "Species Scientific Name" # [5] "Annotation Date" "Sequence Type" # [7] "Sequence Source" "Target Description" # [9] "Representative Public ID" "Gene Title" # [11] "Gene Symbol" "ENTREZ_GENE_ID" # [13] "RefSeq Transcript ID" "SGD accession number" # [15] "Gene Ontology Biological Process" "Gene Ontology Cellular Component" # [17] "Gene Ontology Molecular Function" head(fData(e)$"Gene Symbol") # [1] JIP3 MID2 RPS25B NUP2 # 4869 Levels: ACO1 ARV1 ATP14 BOP2 CDA1 CDA2 CDC25 CDC3 CDD1 CTS1 DBP9 DCS1 ECI1 ECM38 ERF2 EST1 ... Il4 head(rownames(e)) # [1] "10000_at" "10001_at" "10002_i_at" "10003_f_at" "10004_at" "10005_at"
此外还有一些对该研究数据的注释信息,可使用experimentData和annotation函数提取。
experimentData(e) # Experiment data # Experimenter name: # Laboratory: # Contact information: # Title: # URL: # PMIDs: # No abstract available. annotation(e) # [1] "GPL90"
SummarizedExpression演示使用“parathyroidSE”这个数据。
library(parathyroidSE) data(parathyroidGenesSE) se <- parathyroidGenesSE se # class: SummarizedExperiment # dim: 63193 27 # exptData(1): MIAME # assays(1): counts # rownames(63193): ENSG00000000003 ENSG00000000005 ... LRG_98 LRG_99 # rowData metadata column names(0): # colnames: NULL # colData names(8): run experiment ... study sample dim(se) # [1] 63193 27
assay函数提取se的表达矩阵,单元的值为reads的count,类似于expr,包含样本的信息。
assay(se)[1:3,1:3] # [,1] [,2] [,3] # ENSG00000000003 792 1064 444 # ENSG00000000005 4 1 2 # ENSG00000000419 294 282 164 dim(assay(se)) # [1] 63193 27
colData函数类似pData,在此提取外显子的信息,rowData类似fData。
colData(se)[1:3,1:6] # DataFrame with 3 rows and 6 columns # run experiment patient treatment time submission # <character> <factor> <factor> <factor> <factor> <factor> # 1 SRR479052 SRX140503 1 Control 24h SRA051611 # 2 SRR479053 SRX140504 1 Control 48h SRA051611 # 3 SRR479054 SRX140505 1 DPN 24h SRA051611 dim(colData(se)) # [1] 27 8 names(colData(se)) # [1] "run" "experiment" "patient" "treatment" "time" "submission" "study" # [8] "sample" colData(se)$treatment # [1] Control Control DPN DPN OHT OHT Control Control DPN DPN DPN OHT # [13] OHT OHT Control Control DPN DPN OHT OHT Control DPN DPN DPN # [25] OHT OHT OHT # Levels: Control DPN OHT
rowData类似于pData,提取基因的信息,是一个GRangesList的类,每个元素代表一个基因,为一个GRanges的类,包含在内的是外显子的集合。
rowData(se)[1] # GRangesList object of length 1: # $ENSG00000000003 # GRanges object with 17 ranges and 2 metadata columns: # seqnames ranges strand | exon_id exon_name # <Rle> <IRanges> <Rle> | <integer> <character> # [1] X [99883667, 99884983] - | 664095 ENSE00001459322 # [2] X [99885756, 99885863] - | 664096 ENSE00000868868 # [3] X [99887482, 99887565] - | 664097 ENSE00000401072 # [4] X [99887538, 99887565] - | 664098 ENSE00001849132 # [5] X [99888402, 99888536] - | 664099 ENSE00003554016 # ... ... ... ... ... ... ... # [13] X [99890555, 99890743] - | 664106 ENSE00003512331 # [14] X [99891188, 99891686] - | 664108 ENSE00001886883 # [15] X [99891605, 99891803] - | 664109 ENSE00001855382 # [16] X [99891790, 99892101] - | 664110 ENSE00001863395 # [17] X [99894942, 99894988] - | 664111 ENSE00001828996 # # ------- # seqinfo: 580 sequences (1 circular) from an unspecified genome class(rowData(se)) # [1] "GRangesList" # attr(,"package") # [1] "GenomicRanges" length(rowData(se)) # [1] 63193 head(rownames(se)) # [1] "ENSG00000000003" "ENSG00000000005" "ENSG00000000419" "ENSG00000000457" "ENSG00000000460" # [6] "ENSG00000000938" metadata(rowData(se)) # $genomeInfo # $genomeInfo$`Db type` # [1] "TranscriptDb" # # $genomeInfo$`Supporting package` # [1] "GenomicFeatures" # # $genomeInfo$`Data source` # [1] "BioMart" # # $genomeInfo$Organism # [1] "Homo sapiens" # # $genomeInfo$`Resource URL` # [1] "www.biomart.org:80" # # $genomeInfo$`BioMart database` # [1] "ensembl" # # $genomeInfo$`BioMart database version` # [1] "ENSEMBL GENES 72 (SANGER UK)" # # $genomeInfo$`BioMart dataset` # [1] "hsapiens_gene_ensembl" # # $genomeInfo$`BioMart dataset description` # [1] "Homo sapiens genes (GRCh37.p11)" # # $genomeInfo$`BioMart dataset version` # [1] "GRCh37.p11" # # $genomeInfo$`Full dataset` # [1] "yes" # # $genomeInfo$`miRBase build ID` # [1] NA # # $genomeInfo$transcript_nrow # [1] "213140" # # $genomeInfo$exon_nrow # [1] "737783" # # $genomeInfo$cds_nrow # [1] "531154" # # $genomeInfo$`Db created by` # [1] "GenomicFeatures package from Bioconductor" # # $genomeInfo$`Creation time` # [1] "2013-07-30 17:30:25 +0200 (Tue, 30 Jul 2013)" # # $genomeInfo$`GenomicFeatures version at creation time` # [1] "1.13.21" # # $genomeInfo$`RSQLite version at creation time` # [1] "0.11.4" # # $genomeInfo$DBSCHEMAVERSION # [1] "1.0"
exptData和abstract函数能提取关于研究的注释信息。
exptData(se)$MIAME # Experiment data # Experimenter name: Felix Haglund # Laboratory: Science for Life Laboratory Stockholm # Contact information: Mikael Huss # Title: DPN and Tamoxifen treatments of parathyroid adenoma cells # URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37211 # PMIDs: 23024189 # # Abstract: A 251 word abstract is available. Use ‘abstract‘ method. abstract(exptData(se)$MIAME)
ExpressionSet和SummarizedExpression的构建
以“GSE5859Subset”这个数据集为例。
library(devtools)
install_github("genomicsclass/GSE5859Subset")
library(GSE5859Subset)
data(GSE5859Subset)
dim(geneExpression)
dim(sampleInfo)
dim(geneAnnotation)
在每次构建ExpressionSet对象的时候,必须要确保sampleInfo的row对应的是geneExpression的column,geneAnnotation的row对应的是geneExpression的row,将sampleInfo和geneAnnotation的rownames与ExpressionSet的row和column一一对应。如果sampleInfo和geneAnnotation原格式为Df类,使用AnnotatedDataFrame函数转换成AnnotatedDataFrame类。使用ExpressionSet函数构建ExpressionSet对象。
identical(colnames(geneExpression),sampleInfo$filename) identical(rownames(geneExpression),geneAnnotation$PROBEID) library(Biobase) rownames(sampleInfo) <- sampleInfo$filename rownames(geneAnnotation) <- geneAnnotation$PROBEID eset <- ExpressionSet(assayData = geneExpression, phenoData = AnnotatedDataFrame(sampleInfo), featureData = AnnotatedDataFrame(geneAnnotation))
接着我们尝试将eset改建为一个SummarizedExpression对象,基本的步骤同ExpressionSet,不同的是SummarizedExpression的row是记录关于基因位置的信息。
首先加载Homo.sapiens包并使用gene函数提取Homo.sapiens的annotation信息(“GSE5859”这组数据的种属是Homo sapiens)。
library(Homo.sapiens) genes <- genes(Homo.sapiens)
然后加载hgfocus.db,由于eset的feature是“PROBEID”,我们将把它作为key使用select函数从hgfocus.db中提取关联的“ENTREZID”,并用“ENTREZID”作为索引从genes中提取基因位置信息。
library(hgfocus.db) map <- select(hgfocus.db, keys=featureNames(eset), columns="ENTREZID",keytype="PROBEID") head(map) # PROBEID ENTREZID # 1 1007_s_at 780 # 2 1007_s_at 100616237 # 3 1053_at 5982 # 4 117_at 3310 # 5 121_at 7849 # 6 1255_g_at 2978
select函数会提示“‘select‘ resulted in 1:many mapping between keys and return rows”的警告信息,说明有不止一个“ENTREZID”和“PROBEID”匹配,比如“1007_s_at”就同时有两个匹配:780和100616237。
我们使用match函数自动提取第一个“ENTREZID”匹配,再用“ENTREZID”匹配在gene的metadata中的“GENEID”,移除没有配对成功的ID(NA值)。
index1 <- match(featureNames(eset),map[,1]) index2 <- match(map[index1,2], as.character(mcols(genes)$GENEID)) index3 <- which(!is.na(index2)) index2 <- index2[index3]
构建新的匹配成功的子集,并构建SummarizedExperiment。
genes <- genes[index2,] neweset <- eset[index3,] se <- SummarizedExperiment(assays=exprs(neweset), rowData=genes, colData=DataFrame(pData(neweset)))
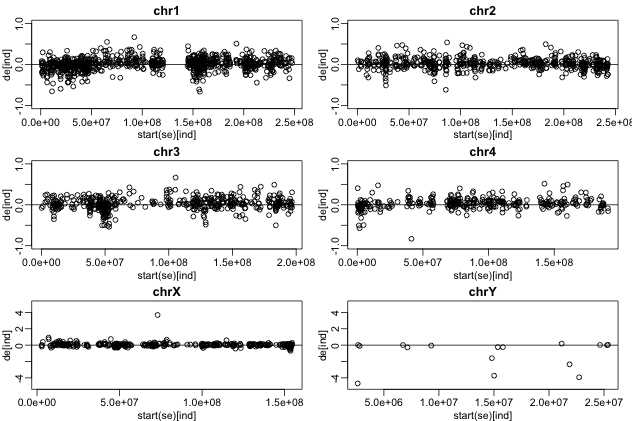
最后,我们还可以非常漂亮地画出不同染色体上差异表达情况。
se = se[order(granges(se)),]
ind = se$group==1
de = rowMeans( assay(se)[,ind])-rowMeans( assay(se)[,!ind])
chrs = unique( seqnames(se))
library(rafalib)
mypar2(3,2)
for(i in c(1:4)){
ind = which(seqnames( se) == chrs[i])
plot(start(se)[ind], de[ind], ylim=c(-1,1),main=as.character(chrs[i]))
abline(h=0)
}
##now X and Y
for(i in 23:24){
ind = which(seqnames( se) == chrs[i])
##note we use different ylims
plot(start(se)[ind], de[ind], ylim=c(-5,5),main=as.character(chrs[i]))
abline(h=0)
}
标签:
原文地址:http://www.cnblogs.com/nutastray/p/4438024.html